真菌毒素(Mycotoxin)是由丝状真菌(如镰刀菌属、曲霉属和青霉属等)产生的次级代谢产物。目前已知的真菌代谢产物至少有400 多种,其中至少有30 多种对动物和人类具有潜在毒性及危害,包括能够产生致癌、致突变、致畸、免疫抑制、内分泌干扰和肝肾功能异常等作用,急性毒性还可能直接造成人类及动物的死亡[1-2]。虽然目前在农业生产的各个环节,应用多种手段来控制真菌的污染和繁殖,但是在农作物生产过程中产毒真菌及真菌毒素的污染问题仍是全球高发性的食品安全问题。北美与东亚是真菌毒素污染的高发地区,有调查显示,全球72 个国家的18 757 种农产品被真菌毒素所污染。在我国,脱氧雪腐镰刀菌烯醇(Deoxynivalenol,DON)、玉米赤霉烯酮(Zearalenone,ZEA)、黄曲霉毒素(Aflatoxins,AFs)、伏马毒素(Fumonisins,FBs)、赭曲霉毒素A(Ochratoxin A,OTA)、展青霉素(Patulin,PAT)等常见真菌毒素在农产品中的检出率均超过25%[3]。真菌毒素污染主要发生在田间种植阶段与采后贮藏阶段,导致植株病害及动物食用受污染的饲料,从而引起疾病或死亡,直接导致农作物产量减少与牲畜生产力的下降。在食品加工环节,由于真菌毒素复杂的分子结构及理化性质,因此传统的加工方式对其破坏程度十分有限。在农产品的市场流通及国际贸易中,因真菌毒素污染造成商品无法出口的问题时有发生,给出口国造成了巨大的经济损失[4-5]。欧盟(European Union,EU)、美国食品和药物管理局(Food and Drug Administration,FDA)和其它一些国家已对受真菌毒素影响的商品制定了监管准则和限制。因此,无论是从经济还是从食品安全的角度,真菌毒素污染的问题都亟待解决。
一直以来人们都在积极寻找并优化真菌毒素的脱除方法。目前有效脱除农作物中真菌毒素的方法主要分为物理法、化学法和生物法三大类。物理法和化学法具有成本高,效率低,安全性不足,可能破坏食物中必需的营养物质,适口性降低等缺点,脱毒效果并不理想[6]。而生物脱毒法是利用微生物自身特性或其生长过程中的代谢产物对毒素进行降解、转化或吸附,使毒性较强的物质转变为无毒或低毒性物质,此类方法具有温和、高效、污染小、特异性强等优点[7],目前已发现许多细菌、霉菌和酵母具有高效脱除真菌毒素的能力。开发(微)生物脱毒技术对于提高食品安全性,减少潜在的农业经济损失至关重要[8-9],已成为国内外真菌毒素脱毒方法的研究热点。
1 农产品中真菌毒素概述
自然界中现存数以千计的真菌毒素,其中与农产品相关的约有几百种,主要由曲霉属(Aspergillus spp.)、青霉属(Penicillium spp.)、镰刀菌属(Fusarium spp.)等真菌代谢产生,这些产毒真菌在自然界中无处不在,广泛存在于土壤、空气和植物中,其产生毒素的化学结构也非常多样,具有污染食物和动物饲料的能力[4,10]。小麦、玉米、大麦等谷物及豆类、坚果、咖啡、水果等农产品的污染情况尤为严重,极大地影响了食品品质和经济价值,危害人类健康[11]。
目前在农产品中污染最严重的真菌毒素分别为AFs、OTA、FBs、ZEA、PAT、DON 等,其中以AFB1毒性最强,被世界国际癌症研究机构(International agency for research on cancer,IARC)归类为1 类致癌物(对人具有致癌性),此外,OTA 和FB1 也被IARC 列为2B 类(对人可能致癌)致癌物[12]。农产品中常见各类真菌毒素的代表性结构、主要污染对象、毒性作用和病理表现以及我国现行的限量标准见表1。
表1 农产品中主要的真菌毒素及其污染对象
Table 1 Several main types of mycotoxins in agricultural products

(续表1)
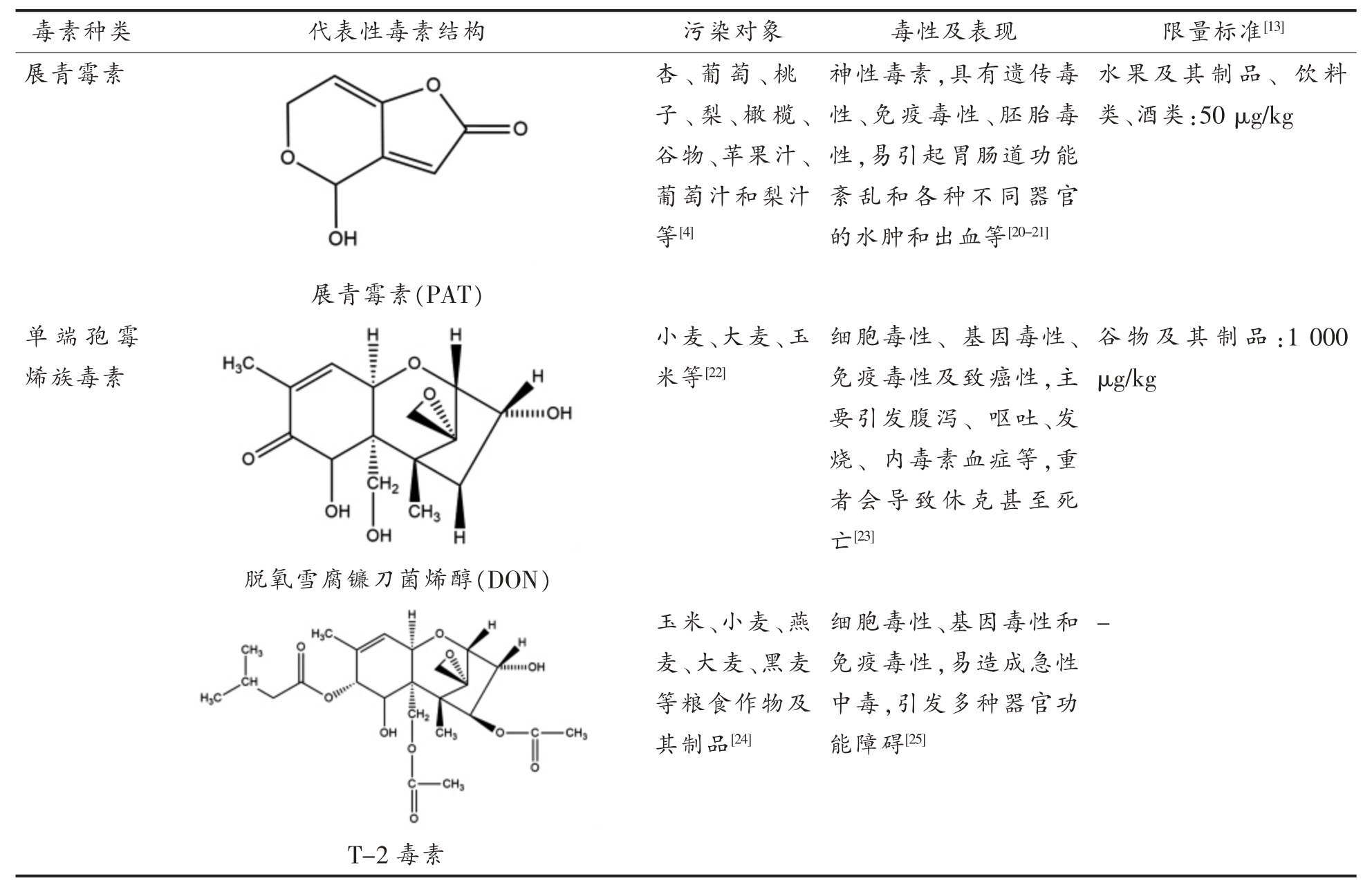
注:表中“-”代表我国暂未对该种毒素制定限量标准。
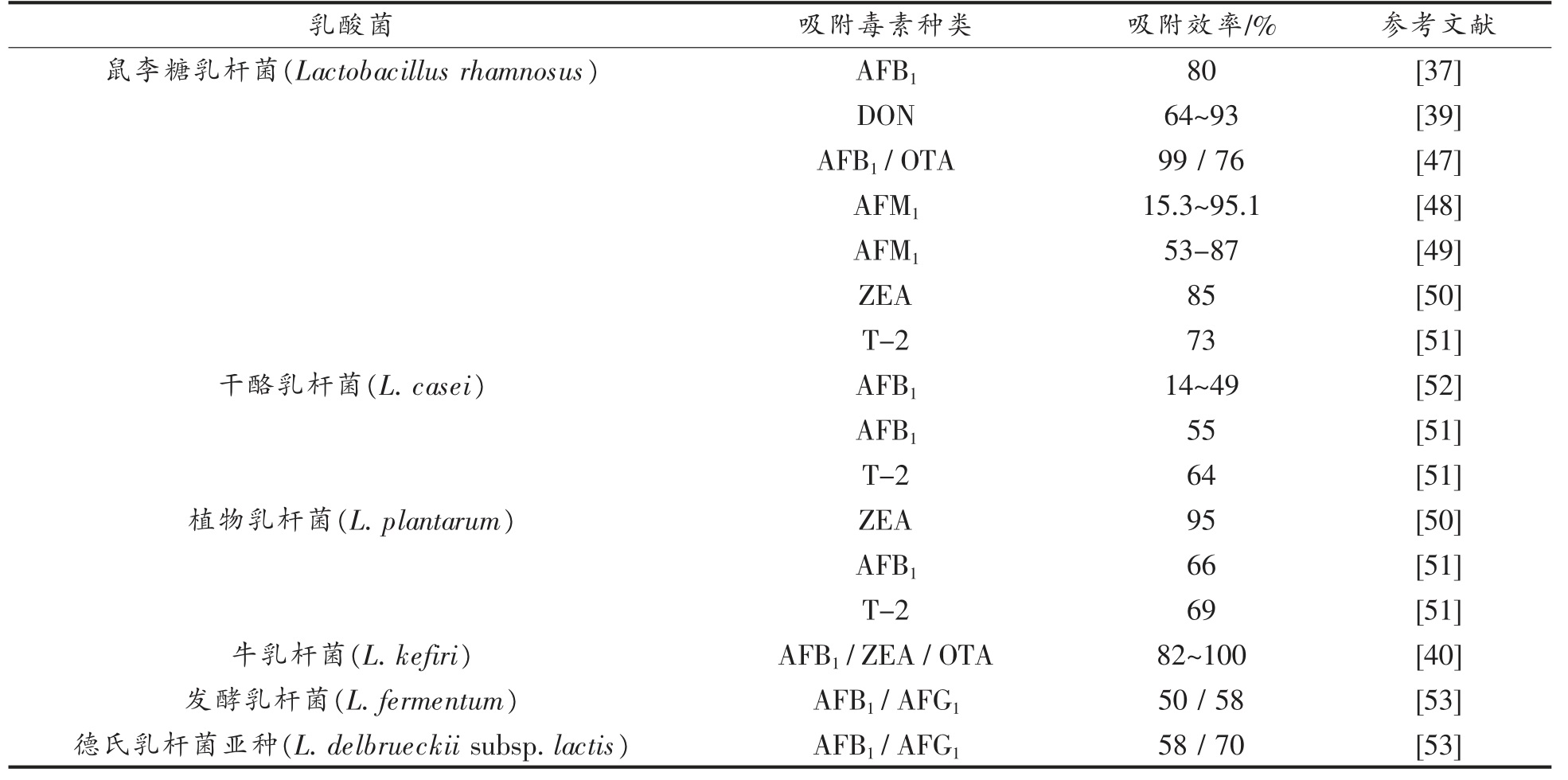
表2 乳酸菌对真菌毒素的吸附作用
Table 2 The adsorption of lactic acid bacteria on mycotoxins
2 真菌毒素的微生物脱除方法
早期的真菌毒素脱除主要依靠物理法和化学法,然而极易导致食品的营养价值损失和生物安全风险,且效果十分有限[5,26]。近年来,农产品中真菌毒素的控制研究越来越多地聚焦在微生物的脱毒作用,因该法具有接受率较高,脱毒效率高,稳定性强,专一性强,安全性强,成本较低等优势,逐渐成为控制真菌毒素污染的主流方法[27-28]。目前诸多研究证明微生物脱除真菌毒素的机制分别是:①选择性膜吸附作用,这主要与菌体细胞壁有关,研究主要集中在乳酸菌和酵母菌的膜吸附;②选择性降解作用,主要利用微生物的代谢产物对毒素进行选择性的体内或体外的降解与转化,使其成为无毒或低毒化合物[29],主要涉及红球菌、芽孢杆菌、假单胞菌以及酵母等微生物。
2.1 微生物菌体吸附法
微生物吸附是利用微生物菌体本身的特性,菌体细胞壁能够以非共价键的形式结合毒素分子[30],形成菌体-毒素复合物而发挥脱毒作用[31]。菌体-毒素复合物结构相对稳定,经过反复水洗后,只有极少数复合物会发生解吸附现象,是一种有限的可逆性过程[32]。由于这种结合方式是物理作用,因此不仅活菌具有吸附毒素的作用,失活的菌体同样也能够发挥很好的毒素脱除作用。乳酸菌和酵母菌等微生物由于其完整的细胞壁结构而表现出对真菌毒素的高效吸附能力[33-35],二者对于真菌毒素的吸附作用如图1 所示。
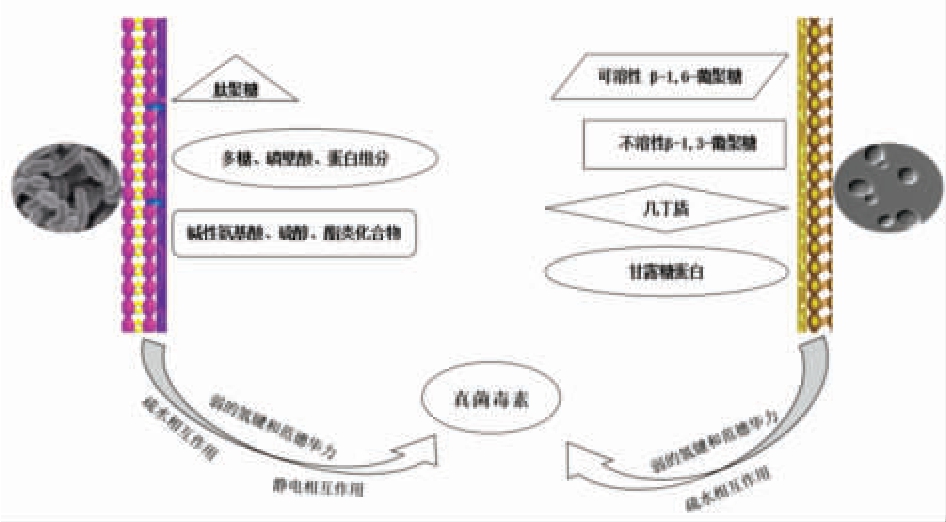
图1 乳酸菌与酵母对真菌毒素的吸附作用示意图
Fig.1 Schematic diagram of the adsorption of lactic acid bacteria and yeast on mycotoxins
2.1.1 乳酸菌的吸附作用 乳酸菌(Lactic acid bacteria)在许多食物中是天然存在的,几百年来一直是人类饮食的一部分,已经是一种全世界公认安全的食用有机体[36]。它是目前用于吸附真菌毒素研究最为广泛的一类微生物。早在1998 年,El-Nezami 等[37]就对乳酸菌在体外结合AFB1 的能力进行了评估,证明AFB1 能够快速与鼠李糖乳杆菌(Lactobacillus rhamnosus)相结合,去除率高达80%,Hernandez-Mendoza 等[32]在乳酸菌的体内研究中也发现其对AFB1 能高效、快速的吸附。尽管有研究证明这种脱除效果是可逆的,乳酸菌对AFB1 的脱除效果仅依赖于菌体细胞表面的结合,而并非酶降解[38],但这并没有影响乳酸菌吸附脱除真菌毒素成为该研究领域长期关注的热点。
乳酸菌对DON、ZEA、OTA 等真菌毒素同样具有吸附脱除的效果。鼠李糖乳杆菌(L.rhamnosus)被证明除了对AFB1 有脱除效果外,对DON 的脱除效果同样喜人,与20 μg/mL 的DON 共同培养1 h 后,吸附率就能够达到64%~93%,且活细胞与灭活细胞对其吸附效果没有显著差异(P>0.05)[39],足以证明这种吸附作用的优异。最新研究表明,牛乳杆菌(L.kefiri)具有很高的吸附活性,能够在牛奶中与其它微生物共同作用,对AFB1、OTA、ZEA 多种真菌毒素的吸附率达到82%~100%[40]。
近年来对乳酸菌吸附真菌毒素的认知不断深入,最初研究认为细胞壁结构中的肽聚糖参与了吸附的过程[41-42],随着研究的深入,发现细胞壁中的磷壁酸、多糖及蛋白质才是乳酸菌吸附真菌毒素的关键成分[32,43],更为细致的研究显示,乳酸菌细胞壁中碱性氨基酸、硫醇和酯类化合物才是对PAT 吸附最为重要的因素[44]。细胞壁成分与毒素之间作用力引起了研究者的关注[43,45],有研究发现两者之间存在静电相互作用与疏水相互作用,而疏水作用更重要。灭活的乳酸菌细胞对于真菌毒素的吸附效果甚至优于活细胞,Niderkorn 等[46]认为,这是由疏水相互作用导致,热处理或酸处理时,细胞壁肽聚糖糖苷键或肽键断裂,使肽聚糖结构变薄,孔径增大,疏水作用明显提高,增强了菌体对真菌毒素的吸附能力。
2.1.2 酵母的吸附作用 酿酒酵母(Saccharomyces cerevisiae)是与人类生活关系最密切的酵母菌,它对多种毒素都具有显著的吸附作用[51]。Bejaoui 等[54]发现酿酒酵母能够脱除葡萄汁中的OTA,将酿酒酵母高温灭活后,其OTA 的吸附率高达90%,显著高于活细胞(对OTA 的吸率仅为35%)[55]。失活的酿酒酵母同样能够降低苹果汁中PAT 的含量,最大吸附量可以达到70.28%[56]。类似的结果在AFB1 的吸附试验同样被观察到,失活的酿酒酵母在5 min 之内即可结合99.3%的AFB1[57]。上述的研究证明酵母细胞对真菌毒素的脱除机制是一种物理性结合,而细胞密度对吸附率起重要作用[55]。
除酿酒酵母外,假丝酵母属(Candida spp.)、克勒克酵母属(Kloeckera spp.)、毕赤酵母属(Pichia spp.)、裂殖酵母属(Schizosaccharomyces spp.)和红酵母属(Rhodotorula spp.)也都具有吸附OTA 和PAT 的能力[58-59]。
在吸附真菌毒素方面,酵母菌的优异潜力更多体现在它能够同时吸附多种真菌毒素。农产品经常会不同程度地被AFs、FB1、ZEA、DON 等多种真菌毒素同时污染。酿酒酵母能同时快速的吸附AFB1 和FB1 两种毒素,且彼此不会竞争酵母细胞壁上的结合位点[34]。美极梅奇酵母(Metschnikowia pulcherrima)、发酵地霉酵母(Geotrichum fermentans)和马克思克鲁维酵母(Kluyveromyces marxianus)等同样对AFs、ZEA、DON 具有良好的吸附效果,甚至能够完全脱除AFs[60]。从发酵乳制品中分离出的瑟氏哈萨克斯坦酵母(Kazachstania servazzii)也能够同时吸附AFB1、ZEA 和OTA[40]。
酵母菌对真菌毒素的吸附机理研究,目前大多数侧重于酵母细胞壁结构与形态、细胞壁的组成成分以及上述二者之间的相互作用[35]。Luo 等[59]研究了4 株酵母细胞对PAT 的吸附能力,发现具有更致密、更有厚度的细胞壁三维网状结构,对PAT 具有更好的吸附能力,而一旦破坏了这种三维网络结构会显著降低这种吸附能力。Pereyra 等[61]的研究发现酵母细胞壁的厚度对AFB1 吸附具有重要影响,这进一步验证、丰富了Luo 等[59]对细胞壁结构影响真菌毒素吸附性能的认知,引导研究人员将酵母菌对真菌毒素的吸附作用机制聚焦于酵母菌细胞壁结构与组成。酵母细胞壁的三维网络结构主要由几丁质、β-1,6-葡聚糖和β-1,3-葡聚糖侧链连接甘露蛋白组成[62]。上述细胞壁组成成分被证明对毒素吸附起重要作用[59,63-65]。与单纯研究酵母菌细胞壁结构、细胞壁组成成分对真菌毒素的吸附作用相比,近年来越来越多的研究发现,二者的相互作用能够更好的解释酵母菌的吸附作用[35],最近一项研究发现,一定浓度的外源Ca2+作用能上调β-1,3-葡聚糖酶和β-1,3-糖基转移酶的基因表达,通过增强关键催化酶的活性,增加β-1,3-葡聚糖含量,进而增强酵母细胞壁的厚度,改变细胞壁三维网状结构,增强其对PAT的吸附能力[66]。这项研究为真菌毒素的吸附脱除提供了新的思路,科研人员可以尝试通过基因调控诱导酵母菌细胞壁,形成更易与各类真菌毒素结合的空间构型,使酵母菌成为一种更为全面、有效和安全的真菌毒素生物脱除剂。
2.1.3 其它微生物的吸附作用 除乳酸菌与酵母菌外,一些微生物也被证明具有一定的吸附真菌毒素能力。从发酵大豆中分离出的黑曲霉菌株FS10(Aspergillus niger strain FS10)可以降低玉米浆中ZEA 的水平,其菌体能够有效吸附液体培养基中89.56%的ZEA,以及玉米浆原液中60.01%的ZEA[67]。脂环酸杆菌(Alicyclobacillus spp.)的灭活菌株能够有效吸附苹果汁中的PAT,且对苹果汁的理化性质没有特殊影响[68]。丙酸杆菌(Propionibacterium spp.)在酸化的MRS 肉汤(pH 4.0)中能有效吸附FB1 和FB2[69]。此外,粪肠球菌(Enterococcus faecium)[70]、地衣芽孢杆菌CK1(Bacillus licheniformis strain CK1)[71]等微生物都有一定吸附真菌毒素的能力。
2.2 微生物降解法
利用微生物的代谢作用及相关酶可以使真菌毒素降解或转化,破坏其毒性结构,使其成为无毒或毒性较低的化合物,该过程一般通过乙酰化、糖基化、环裂解、水解、脱氨和脱羧作用实现[5]。相比于其它脱毒方法,微生物降解不会吸附食品中的营养物质,也不存在微生物吸附的可逆性问题,是脱除农产品中真菌毒素的理想手段[27-28,72-73]。利用微生物降解真菌毒素的研究始自20 世纪60 年代末,Ciegler 等[74]发现1 株棒状诺卡氏菌(早前被称为橙色黄杆菌)能在28 ℃下降解液体培养基中74%的AFB1,这是第1 株具有降解真菌毒素作用的微生物。此后,有关微生物对真菌毒素降解作用的研究不断涌现。研究主要集中在2 个方向:一是广泛寻找降解真菌毒素的微生物,不断提高其降解效果;二是探究微生物降解真菌毒素的机制。虽然降解机制研究是今后开发降解真菌毒素酶制剂的基础,但目前研究仍集中于寻找适宜的降解微生物资源。事实证明,此类微生物资源相对集中在红球菌、芽孢杆菌、假单胞菌等部分细菌,以及部分酵母菌和霉菌。
2.2.1 红球菌的降解作用 红球菌(Rhodococcus)[75]、链霉菌(Streptomyces)[76-77]、诺卡氏菌(Nocardia)[74,78]、短杆菌(Brevundimonas)[79-80]和分枝杆菌(Mycobacterium)[81-82]属于放线菌科,是土壤中最重要的原核菌群之一[77],能降解不同种类的真菌毒素,其中以红球菌属的降解能力最强,表3 总结了近年来红球菌降解真菌毒素的研究成果。
表3 红球菌属对真菌毒素的降解作用
Table 3 Degradation of Rhodococcus to mycotoxins
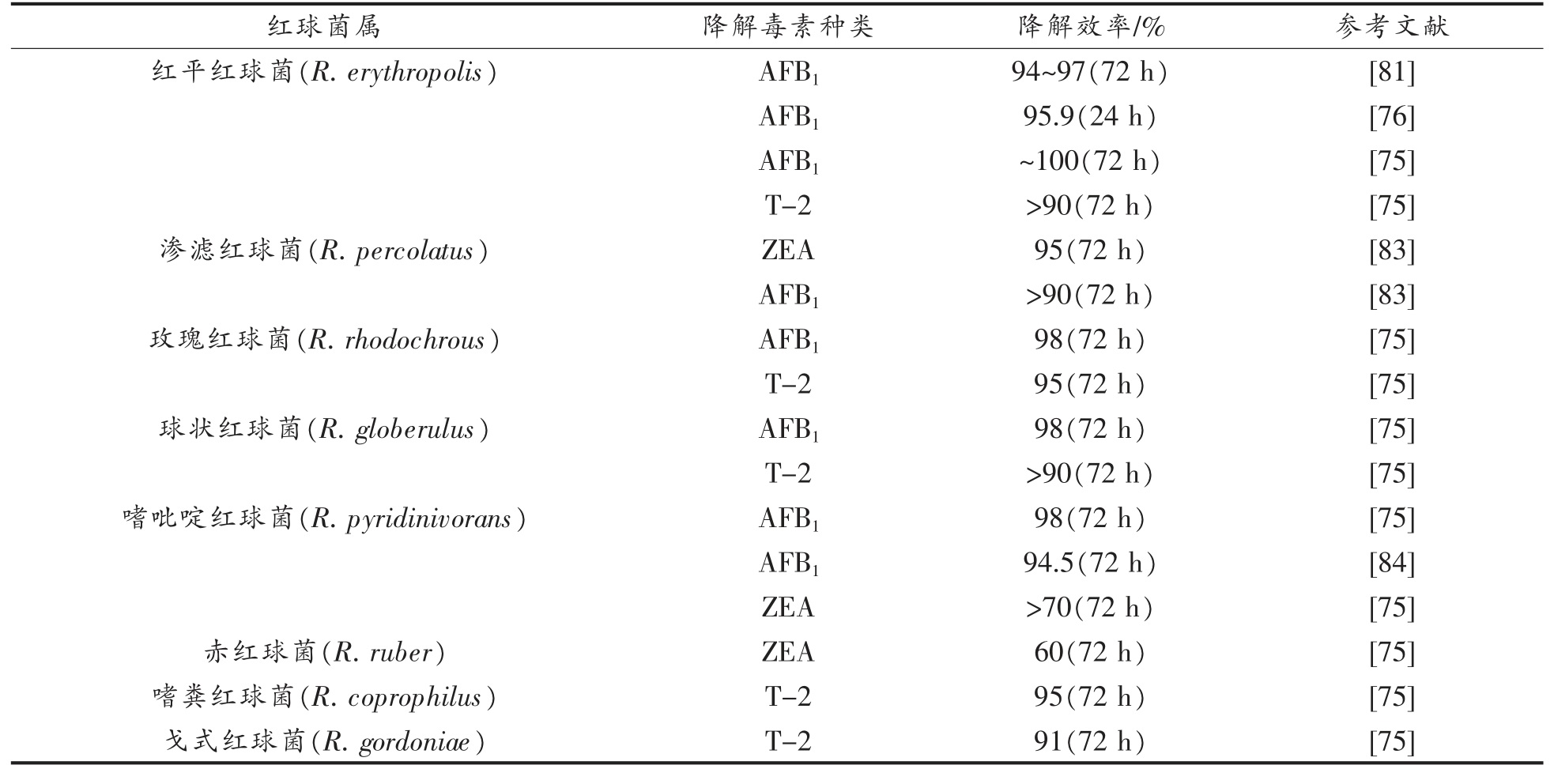
从表3 可以看出,红球菌对芳香族毒素(包括AFB1、ZEA、T-2 等)的降解作用尤为突出,对AFB1 的降解效率都能达到90%以上。红平红球菌对培养基中AFB1 的降解效率高达100%,对难以降解的T-2 毒素,也具有90%以上降解率[75]。红球菌属微生物对此类真菌毒素的降解作用在Risa等[83]的研究中得到了进一步的肯定,该研究发现了18 种红球菌对培养基中的AFB1 具有降解活性,且降解率高达90%以上,证明该类微生物在降解真菌毒素方面的资源潜力和不凡的水平。
针对真菌毒素的微生物降解作用,不仅应关注降解效率,更应该关注降解产物的毒性及安全性。有研究以红平红球菌ATCC 4277(R.erythropolis ATCC 4277)为例,在30 ℃下孵育24 h后,AFB1 的降解率达到95.9%,进一步检测其降解产物后推测,AFB1 被降解成为无毒芳香族化合物(图2 所示),其结构类似脂肪酸代谢和糖酵解的中间产物[76]。然而这并非是所有红平红球菌的共性代谢途径,Cserháti 等[75]通过遗传毒性实验评估真菌毒素代谢后产物的安全性,发现18 株有效降解AFB1 的红平红球菌中,有12 株不产生具有遗传毒性的代谢产物,6 株虽然可以降解AFB1,但其产物仍保留了AFB1 的遗传毒性。
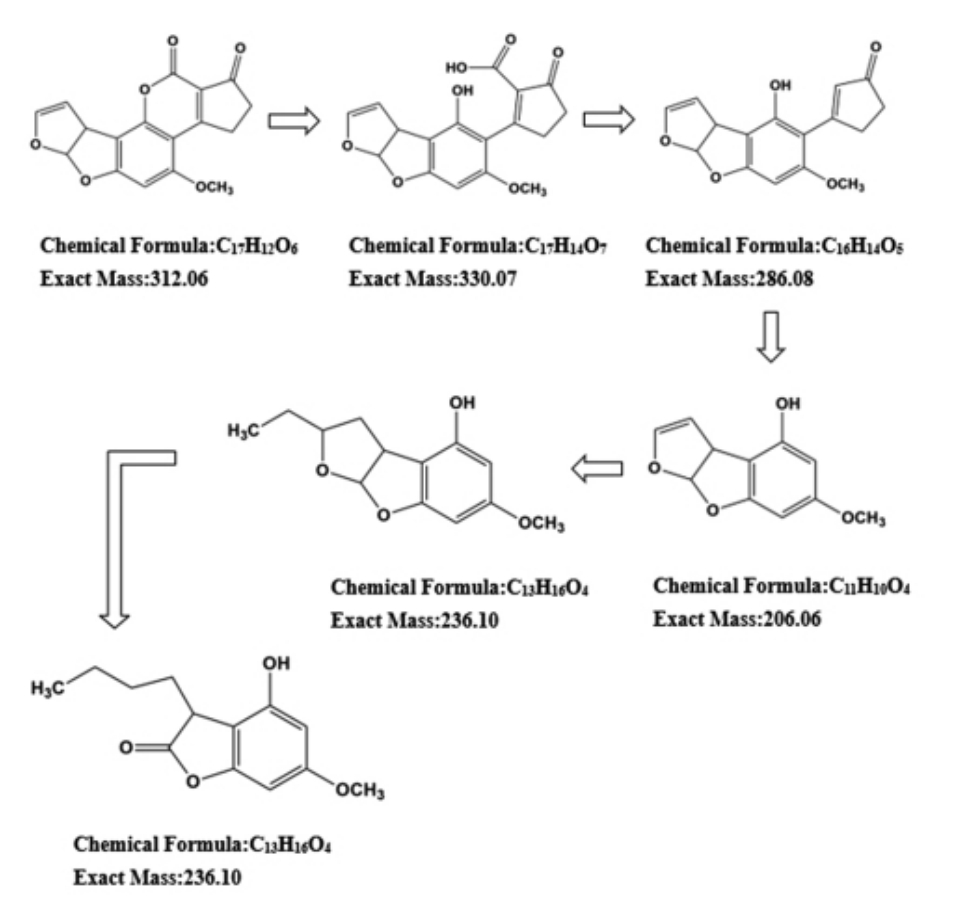
图2 红平红球菌ATCC 4277 对AFB1 的降解途径假说[76]
Fig.2 Hypothesis of the degradation pathway of AFB1 by Rhodococcus erythropolis ATCC 4277[76]
从上述一系列研究报道中不难理解,红球菌在降解真菌毒素中备受瞩目的原因,一方面归功于自身庞大的基因组带来的出色的分解代谢真菌毒素的能力,另一方面则得益于其生物毒性多样化的代谢产物带来的解毒差异性。前者使之获得了降解包括真菌毒素在内的大量有机化合物的能力,颇具工业化应用的潜力[85-87];后者提醒研究者,菌株对真菌毒素的高降解率与其解毒潜力没有明显的相关性[75,83,88],真菌毒素生物降解和脱除是同等重要的研究内容。
2.2.2 芽孢杆菌的降解作用 芽孢杆菌(Bacillus)在降解真菌毒素方面也发挥了重要的作用。它对脱除真菌毒素的有效性一方面体现在通过合成抗菌物质抑制真菌生长及真菌毒素产生[73],另一方面,通过其代谢活动直接降解多种真菌毒素。同时,芽孢杆菌属的一些常见菌种也被认为是益生菌,包括枯草芽孢杆菌(B.subtilis)、地衣芽孢杆菌(B.licheniformis)和纳豆芽孢杆菌(B.natto)等,而这些有益的芽孢杆菌对真菌毒素的脱除也有较好的效果。
如表4 所示,地衣芽孢杆菌(B.licheniformis)对OTA、AFB1、ZEA 和DON 等多种真菌毒素都表现出了高效的降解能力。针对培养基中的OTA,地衣芽孢杆菌CM 21(B.licheniformis CM 21)在48 h 孵育后即可获得92.5%的脱除效果[90];地衣芽孢杆菌CFR1(B.licheniformis CFR1)在与AFB1 共同孵育72 h 后,对其降解率能达到94.7%[89];另一株从土壤中分离出的地衣芽孢杆菌CK1(B.licheniformis CK1)在36 h 后能降解培养基中98%的ZEA[91];而地衣芽孢杆菌NRRL B-50506(B.licheniformis NRRL B-50506)被发现能够在培养基中孵育48 h 后,降解97.2%的DON[92]。
表4 芽孢杆菌对真菌毒素的降解作用
Table 4 Degradation of mycotoxins by Bacillus
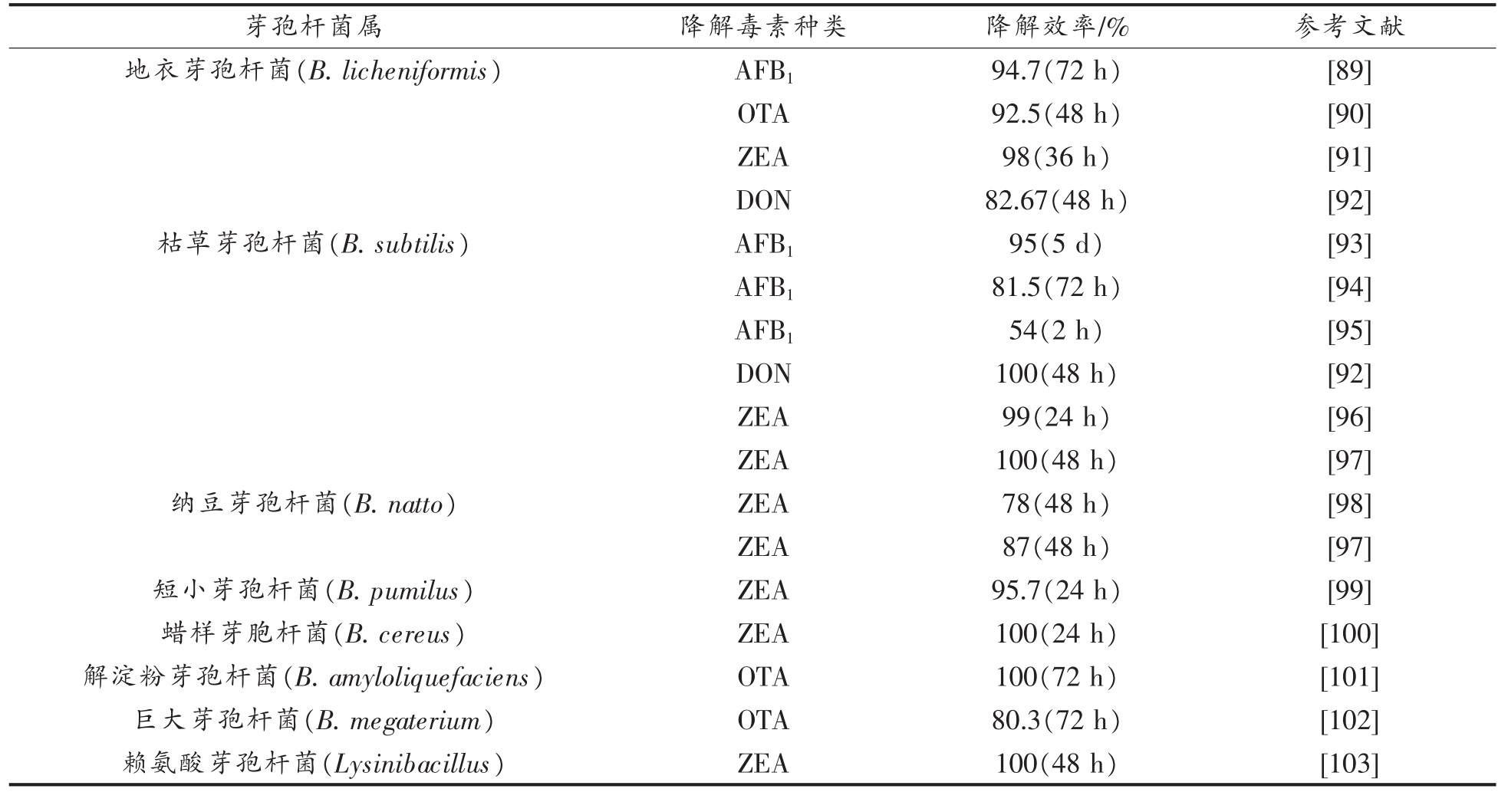
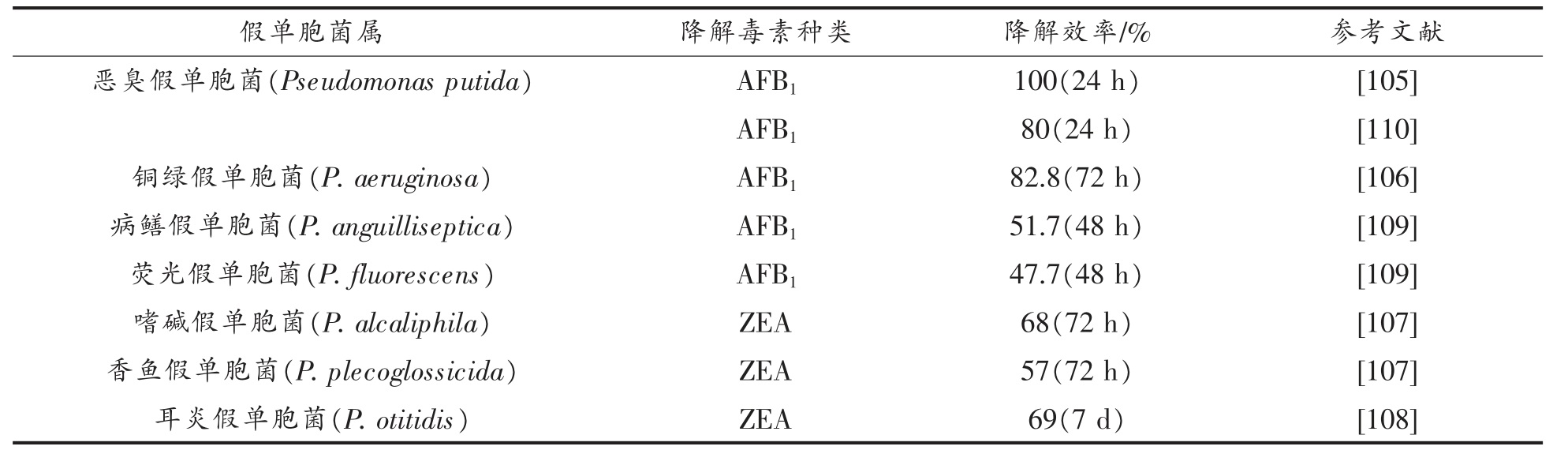
表5 假单胞菌对真菌毒素的降解作用
Table 5 Degradation of mycotoxin by Pseudomonas
尽管芽孢杆菌对多种真菌毒素具有较好的降解效果,但对其研究多集中于ZEA 的降解方面,究其原因,一方面是诸多芽孢杆菌表现出对ZEA的降解活性,包括地衣芽孢杆菌、枯草芽孢杆菌、纳豆芽孢杆菌、短小芽孢杆菌、蜡样芽孢杆菌以及赖氨酸芽孢杆菌等,而且降解效率普遍达到80%~100%[91,97,103],另一方面是由ZEA 自身的结构、代谢产物的多样性决定的。ZEA 是一种非甾体雌激素毒素,进入人体后能被迅速吸收并造成多种毒性,其衍生物同样具有或高或低的雌激素毒性,例如:常见的α-玉米赤霉醇(α-zearalanol)和β-玉米赤霉醇(β-zearalanol)[88],因此降解ZEA 并不代表消除了ZEA 的毒性。因此,目前对芽孢杆菌降解ZEA 的机制多集中在发现其降解产物及降解过程中的生物毒性研究方面。有研究发现ZEA 降解的中间产物——1-(3,5-二羟苯基)-6'-羟基-l'-十一烯-10'-酮,它虽然会迅速分解,但可推测出ZEA 的降解过程经历了内酯环的裂解,随后发生了脱羧反应(如图3 所示),这为后续芽孢杆菌降解ZEA 的代谢产物及机制的研究提供了理论依据[99]。然而在研究赖氨酸芽孢杆菌对ZEA 的代谢产物时,却未检测到任何产物[103],虽然这样的结果可能与检测方法的选择性有关系,但也证明ZEA 代谢产物的复杂性。在ZEA 降解过程中的毒性研究中,Wang 等[100]的研究发现,通过动物体内实验证明了蜡样芽胞杆菌BC7 能够显著改善ZEA 对小鼠造成的肝毒性和生殖毒性,同时可以有效地使ZEA 诱导的肠道菌群扰动正常化,维持小鼠肠道菌群的健康,这使芽孢杆菌成为降解ZEA 的安全选择之一。
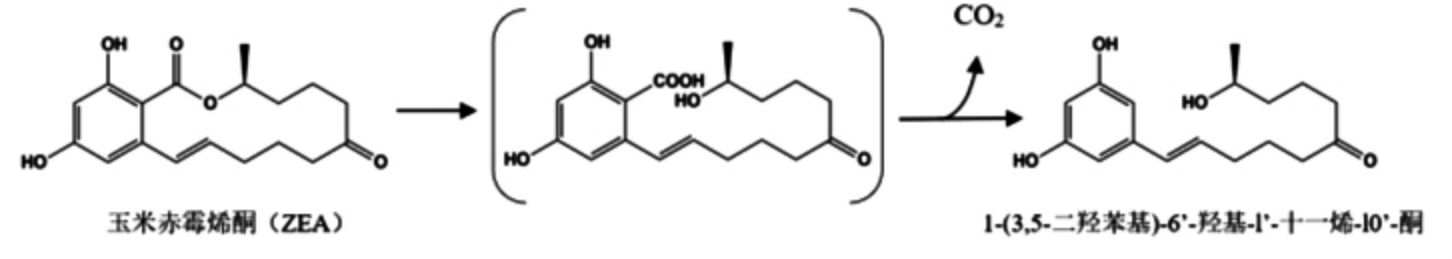
图3 ZEA 降解为1-(3,5-二羟苯基)-6'-羟基-1'-十一烯-10'-酮的机制[99]
Fig.3 The mechanism of ZEA degradation to 1-(3,5-dihydroxyphenyl)-6'-hydroxy-1'-undecene-10'-one[99]
2.2.3 假单胞菌的降解作用 假单胞菌(Pseudomonas)作为一种土壤细菌,被广泛应用于降解各种环境污染物[104],近年来不断发现其具有降解AFB1、ZEA 等真菌毒素的作用[105-109]。
其中最有效的降解菌株是从甘蔗中分离出的恶臭假单胞菌(Pseudomonas putida),在37 ℃孵育24 h 后,能将AFB1 降解至检出限以下,与上文提到的红平红球菌有所不同,在恶臭假单胞菌的代谢过程中,AFB1 分子通过呋喃、内酯环的修饰和环戊烯酮环的丢失,转化为结构不同且毒性较小的新化合物(AFD1、AFD2 和AFD3,如图4 所示)[105],而前者代谢中AFB1 被降解成为无毒芳香族化合物(图2 所示)。在最近关于恶臭假单胞菌对AFB1的降解机制研究中发现,无细胞的培养上清液和细胞裂解物均能够有效减少AFB1,而热灭活的细胞裂解物降解率极低,推测这种降解可能是一个酶促过程[110]。同样的,Song 等[111]也在研究中发现铜绿假单胞菌对AFB1 的降解可能是由于酶的作用,他们进一步从铜绿假单胞菌中分离纯化出了能够降解AFB1 的酶(PADE)并证明了其活性。
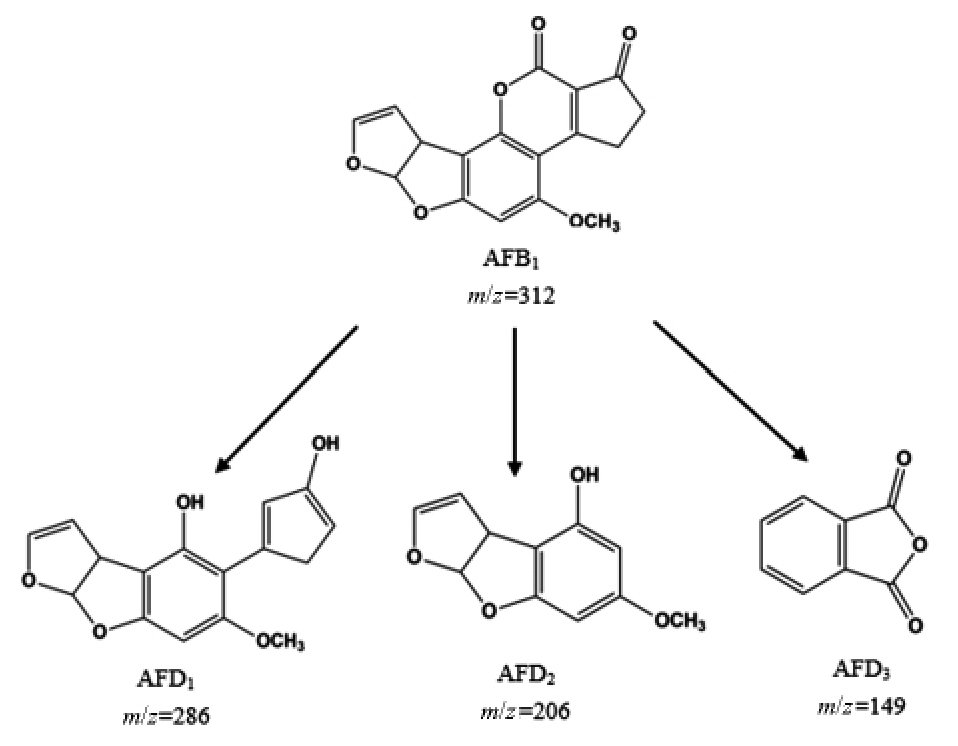
图4 恶臭假单胞菌对AFB1 的降解机理[105]
Fig.4 Degradation mechanism of AFB1 by Pseudomonas putida[105]
2.2.4 酵母菌的降解作用 本文前述,酵母菌通过自身的细胞壁结构,吸附去除展青霉素(PAT),进一步研究证明,酵母菌还可以通过代谢活动降解PAT,这些研究成果进一步使酵母菌在PAT 脱除中成为一种安全、高效的选择。Castoria 等[112]研究了一种科特维乐红酵母LS11(Rhodosporidium kratochvilovae LS11),在培养基中孵育72 h 后,PAT 的含量仅剩2%左右;另外一株胶红酵母JM19(Rhodotorula mucilaginosa JM19)在缓冲液中孵育21 h 后,即可实现PAT 90%的降解效果[113]。
这样高效的降解效果促使对其降解机理研究不断深入,目前的研究发现酵母菌对其降解主要有2 种途径:1 种途径是酵母将PAT 转化为去氧乙酰丙酸(Desoxypatulinic acid,DPA)(如图5 所示),研究发现,DPA 相比于PAT,对人体淋巴细胞、肝细胞的毒性低得多,这种低毒性是PAT 的内酯环水解和硫醇基反应,以致官能团损失的结果[112-113]。与上述PAT 降解机理不同的是,Dong等[114-115]从海洋酵母中分离出1 株Kodameae ohmeri HYJM34 酵母,可将PAT 代谢为(E)-ascladiol 及其同分异构体(Z)-ascladiol(如图5 所示),该代谢产物的毒性明显低于PAT,与PAT 相比,对细胞凋亡的诱导程度较小,没有显著的抑制细胞增殖的作用。同样的,卡利比克毕赤酵母(Pichia caribbica)对PAT 的代谢也是相同机理[115-117]。

图5 PAT 的2 种降解机制示意图[118]
Fig.5 Schematic diagram of two degradation mechanisms of PAT[118]
上述研究表明,酵母菌对PAT 的降解与脱除具有良好的效果,可以通过深入研究,将其作为生物制剂应用到农产品的真菌毒素污染防治中。
3 结语
农产品中真菌毒素的污染是世界普遍面临的食品安全难题,已经成为严重影响人类健康的风险因子,同时也是限制农产品进出口贸易的关键。对于真菌毒素的脱除,尽管物理法与化学法在早期发挥了一定的作用,但是在环保意识与食品安全意识的驱动下,人们更倾向于采取绿色安全的微生物脱毒方法来应对真菌毒素污染问题。
研究不断发现,不同微生物通过吸附或降解的方式具有脱除真菌毒素的能力或潜力,其中,细菌和酵母菌普遍受到关注和深入研究,不断被证明其具有有效性和高效性。然而这并不意味着其它微生物对真菌毒素没有作用。近年来发现,以木霉(Trichoderma)[119]、根霉(Rhizopus)[119]、曲霉(Aspergillus)[120]等发酵食品中经常检出的霉菌也显示出对AFB1 等真菌毒素不同的降解能力。虽然相比于细菌与酵母菌,这类霉菌繁殖周期长,缺乏广泛性与高效性,但是为微生物脱除真菌毒素提供了更多的选择。
现有研究普遍证明细菌和酵母菌具有广泛性和高效性,然而许多菌株对真菌毒素的代谢产物及代谢机理目前还无法明确,有关研究还在不断深入。这些真菌毒素脱除菌株的不断挖掘,以及其代谢机制的深入研究,将为农产品真菌毒素的脱毒技术带来创新性变革,通过微生物的作用显著提升农产品原料、加工生产过程中以及最终食品的安全性。因此,微生物对农产品中真菌毒素的脱除研究已然成为一种发展趋势,大有可为。
[1] AGRIOPOULOU S,STAMATELOPOULOU E,VARZAKAS T.Advances in occurrence,importance,and mycotoxin control strategies:Prevention and detoxification in foods[J].Foods,2020,9(2):137-184.
[2] PFLIEGLER W P,PUSZTAHELYI T,PÓCSI I.Mycotoxins-prevention and decontamination by yeasts[J].Journal of Basic Microbiology,2015,55(7):805-818.
[3] 兰静,金海涛,赵琳,等.玉米真菌毒素污染与控制技术研究进展[J].农产品质量与安全,2020,5:15-21.LAN J,JIN H T,ZHAO L,et al.Research advance in mycotoxin contamination in maize and its control technology[J].Quality and Safety of Agro-Products,2020,5:15-21.
[4] MURPHY P A,HENDRICH S,LANDGREN C,et al.Food mycotoxins:An update[J].Journal of food science,2006,71(5):R51-R65.
[5] HAQUE M A,WANG Y H,SHEN Z Q,et al.Mycotoxin contamination and control strategy in human,domestic animal and poultry:A review[J].Microbial Pathogenesis,2020,142:104095.
[6] SCHATZMAYR G,ZEHNER F,TÄUBEL M,et al.Microbiologicals for deactivating mycotoxins[J].Molecular Nutrition &Food Research,2006,50(6):543-551.
[7] 冯春露,郭祯祥,郭嘉.玉米的真菌毒素及生物脱毒研究进展[J].粮食与油脂,2018,31(9):14-17.FENG C L,GUO Z X,GUO J.Research progress on mycotoxins and biological detoxification of corn[J].Cereals &Oils,2018,31(9):14-17.
[8] YOUNG J C,ZHOU T,YU H,et al.Degradation of trichothecene mycotoxins by chicken intestinal microbes[J].Food and Chemical Toxicology,2007,45(1):136-143.
[9] LUO Y,LIU X J,LI J K.Updating techniques on controlling mycotoxins-A review[J].Food Control,2018,89:123-132.
[10] ADEGBEYE M J,REDDY P R K,CHILAKA C A,et al.Mycotoxin toxicity and residue in animal products:Prevalence,consumer exposure and reduction strategies-A review[J].Toxicon,2020,177:96-108.
[11] 王龑,王刘庆,刘阳.食品中主要真菌毒素生物合成途径研究进展[J].食品安全质量检测学报,2016(6):2158-2167.WANG Y,WANG L Q,LIU Y.Research advances on biosynthetic pathways of the common mycotoxins in food[J].Journal of Food Safety &Quality,2016(6):2158-2167.
[12] ALSHANNAQ A,YU J H,IRITI M.Occurrence,toxicity,and analysis of major mycotoxins in food[J].International Journal of Environmental Research and Public Health,2017,14(6):632.
[13] 中华人民共和国国家卫生和计划生育委员会,国家食品药品监督管理总局.食品安全国家标准食品中真菌毒素限量:GB 2761-2017[S].北京:中国标准出版社,2017:2-5.National Health and Family Planning Commission of the People's Republic of China,State Administration for Market Regulation.Food Safety National Standard Food Limited Food to Toxins Limited:GB 2761-2017[S].Beijing:China Standard Press,2017:2-5.
[14] REDDY K R N,SPADARO D,GULLINO M L,et al.Potential of two Metschnikowia pulcherrima(yeast)strains for in vitro biodegradation of patulin[J].Journal of food protection,2011,74(1):154-156.
[15] 乔宏兴,姜亚乐,王永芬,等.黄曲霉毒素的危害及其脱毒方法研究进展[J].动物医学进展,2017,38(1):89-93.QIAO H X,JIANG Y L,WANG Y F,et al.Progress on harm and detoxification methods of aflatoxin[J].Progress in Veterinary Medicine,2017,38(1):89-93.
[16] 刘青,邹志飞,余炀炀,等.食品中真菌毒素法规限量标准概述[J].中国酿造,2017,36(1):12-18.LIU Q,ZOU Z F,YU Y Y,et al.Review of regulations of mycotoxins limit standard in foods [J].China Brewing,2017,36(1):12-18.
[17] SCHRENK D,BODIN L,CHIPMAN J K,et al.Risk assessment of ochratoxin A in food[J].EFSA Journal,2020,18(5):e06113.
[18] SUN G J,WANG S K,HU X,et al.Fumonisin B1 contamination of home-grown corn in high-risk areas for esophageal and liver cancer in China[J].Food Additives and Contaminants,2007,24(2):181-185.
[19] 王怡净,张立实.玉米赤霉烯酮毒性研究进展[J].中国食品卫生杂志,2002,14(5):40-43.WANG Y J,ZHANG L S.Research progress on toxicity of zearalenone[J].Chinese Journal of Food Hygiene,2002,14(5):40-43.
[20] WEI C Z,YU L L,QIAO N Z,et al.Progress in the distribution,toxicity,control,and detoxification of patulin:A review[J].Toxicon,2020,184:83-93.
[21] GLASER N,STOPPER H.Patulin:Mechanism of genotoxicity[J].Food &Chemical Toxicology,2012,50(5):1796-1801.
[22] 张紊玮,王艳玲,薛华丽,等.镰刀菌单端孢霉烯族毒素的生物合成及分子调控研究进展[J].食品科学,2019,40(5):267-275.ZHANG X W,WANG Y L,XUE H L,et al.Advances in biosynthesis and regulation of Fusarium Trichothecenes[J].Food Science,2019,40(5):267-275.
[23] 李国林,薛华丽,毕阳,等.脱氧雪腐镰刀菌烯醇的毒性及脱毒研究进展[J].食品工业科技,2013,34(24):380-384.LI G L,XUE H L,BI Y,et al.Research progress in toxicity and detoxification of deoxynivalenol [J].Science and Technology of Food Industry,2013,34(24):380-384.
[24] 邹广迅,张红霞,花日茂.T-2 毒素的毒性效应及致毒机制研究进展[J].生态毒理学报,2011,6(2):121-128.ZOU G X,ZHANG H X,HUA R M.Research progress in toxicological effects and mechanism of T-2 toxin[J].Asian Journal of Ecotoxicology,2011,6(2):121-128.
[25] 吴限鑫,林秋君,郭春景,等.国内外主要粮油产品中真菌毒素限量,检测标准及风险评估现状分析[J].中国粮油学报,2019,34(9):130-138.WU X X,LIN Q J,GUO C J,et al.Analysis of limits,testing standards and risk assessment of mycotoxins in major grain and oil products at home and abroad[J].Journal of the Chinese Cereals and Oils Association,2019,34(9):130-138.
[26] 孙玉凤,金诺,刘佳萌,等.生物毒素的脱毒技术及药物研究进展[J].食品安全质量检测学报,2020,11(12):3958-3964.SUN Y F,JIN N,LIU J M,et al.Research progress on detoxification technology and medicine of biological toxin[J].Journal of Food Safety and Quality,2020,11(12):3958-3964.
[27] JI C,FAN Y,ZHAO L H.Review on biological degradation of aflatoxin,zearalenone and deoxynivalenol[J].Animal Nutrition,2016,2(3):127-133.
[28] ADEBIYI J A,KAYITESI E,ADEBO O A,et al.Food fermentation and mycotoxin detoxification:An African perspective[J].Food Control,2019,106:106731.
[29] HATHOUT A S,ALY S E.Biological detoxification of mycotoxins:A review[J].Annals of Microbiology,2014,64(3):905-919.
[30] 成博伦,赵玉濛,胡锦涛,等.几种常见真菌毒素脱毒方法的研究进展[J].基因组学与应用生物学,2015,34(11):2338-2344.CHENG B L,ZHAO Y M,HU J T,et al.Progress of detoxification for several common mycotoxins[J].Genomics and Applied Biology,2015,34(11):2338-2344.
[31] ADEBO O A,NJOBEH P B,GBASHI S,et al.Review on microbial degradation of aflatoxins [J].Critical Reviews in Food Science and Nutrition,2017,57(15):3208-3217.
[32] HERNANDEZ‐MENDOZA A,GUZMAN‐de‐PEÑA D,GARCIA H S.Key role of teichoic acids on aflatoxin B1 binding by probiotic bacteria [J].Journal of Applied Microbiology,2009,107(2):395-403.
[33] ARMANDO M R,PIZZOLITTO R P,DOGI C A,et al.Adsorption of ochratoxin A and zearalenone by potential probiotic Saccharomyces cerevisiae strains and its relation with cell wall thickness[J].Journal of Applied Microbiology,2012,113(2):256-264.
[34] PIZZOLITTO R P,SALVANO M A,DALCERO A M.Analysis of fumonisin B1 removal by microorganisms in co-occurrence with aflatoxin B1 and the nature of the binding process [J].International Journal of Food Microbiology,2012,156(3):214-221.
[35] LUO Y,LIU X J,YUAN L,et al.Complicated interactions between bio -adsorbents and mycotoxins during mycotoxin adsorption:Current research and future prospects[J].Trends in Food Science &Technology,2020,96:127-134.
[36] OLUWAFEMI F,KUMAR M,BANDYOPADHYAY R,et al.Bio-detoxification of aflatoxin B1 in artificially contaminated maize grains using lactic acid bacteria[J].Toxin Reviews,2010,29(3):115-122.
[37] EL-NEZAMI H,KANKAANPAA P,SALMINEN S,et al.Ability of dairy strains of lactic acid bacteria to bind a common food carcinogen,aflatoxin B1[J].Food and Chemical Toxicology,1998,36(4):321-326.
[38] PELTONEN K,EL-NEZAMI H,HASKARD C,et al.Aflatoxin B1 binding by dairy strains of lactic acid bacteria and bifidobacteria[J].Journal of Dairy Science,2001,84(10):2152-2156.
[39] EL-NEZAMI H S,CHREVATIDIS A,AURIOLA S,et al.Removal of common Fusarium toxins in vitro by strains of Lactobacillus and Propionibacterium[J].Food Additives &Contaminants,2002,19(7):680-686.
[40] TAHEUR F B,FEDHILA K,CHAIEB K,et al.Adsorption of aflatoxin B1,zearalenone and ochratoxin A by microorganisms isolated from Kefir grains[J].International Journal of Food Microbiology,2017,251:1-7.
[41] HASKARD C A,EL-NEZAMI H S,KANKAANPÄÄ P E,et al.Surface binding of aflatoxin B1 by lactic acid bacteria[J].Applied and Environmental Microbiology,2001,67(7):3086-3091.
[42] LAHTINEN § S J,HASKARD C A,OUWEHAND§ A C,et al.Binding of aflatoxin B1 to cell wall components of Lactobacillus rhamnosus strain GG[J].Food additives and Contaminants,2004,21(2):158-164.
[43] HASKARD C,BINNION C,AHOKAS J.Factors affecting the sequestration of aflatoxin by Lactobacillus rhamnosus strain GG[J].Chemico-Biological Interactions,2000,128(1):39-49.
[44] WANG L,WANG Z L,YUAN Y H,et al.Identification of key factors involved in the biosorption of patulin by inactivated lactic acid bacteria(LAB)cells[J].PLoS One,2015,10(11):95-105.
[45] WANG L,YUE T L,YUAN Y H,et al.A new insight into the adsorption mechanism of patulin by the heat-inactive lactic acid bacteria cells[J].Food Control,2015,50:104-110.
[46] NIDERKORN V,MORGAVI D P,PUJOS E,et al.Screening of fermentative bacteria for their ability to bind and biotrans form deoxynivalenol,zearalenone and fumonisins in an in vitro simulated corn silage model[J].Food Additives and Contaminants,2007,24(4):406-415.
[47] TURBIC A,AHOKAS J T,HASKARD C A.Selective in vitro binding of dietary mutagens,individually or in combination,by lactic acid bacteria[J].Food Additives &Contaminants,2002,19(2):144-152.
[48] ABBÈS S,SALAH-ABBÈS J B,SHARAFI H,et al.Ability of Lactobacillus rhamnosus GAF01 to remove AFM1 in vitro and to counteract AFM1 immunotoxicity in vivo[J].Journal of Immunotoxicology,2013,10(3):279-286.
[49] VAHIDIMEHR A,KHIABANI M S,MOKARRAM R R,et al.Saccharomyces cerevisiae and Lactobacillus rhamnosus cell walls immobilized on nanosilica entrapped in alginate as aflatoxin M1 binders[J].International Journal of Biological Macromolecules,2020,164:1080-1086.
[50] ![]() MARKOV K,FRECE J,et al.Adhesion of zearalenone to the surface of lactic acid bacteria cells [J].Croatian Journal of Food Technology,Biotechnology and Nutrition,2012,7:49-52.
MARKOV K,FRECE J,et al.Adhesion of zearalenone to the surface of lactic acid bacteria cells [J].Croatian Journal of Food Technology,Biotechnology and Nutrition,2012,7:49-52.
[51] CHLEBICZ A,![]() K.In vitro detoxification of aflatoxin B1,deoxynivalenol,fumonisins,T-2 toxin and zearalenone by probiotic bacteria from genus Lactobacillus and Saccharomyces cerevisiae yeast[J].Probiotics and Antimicrobial Proteins,2020,12(1):289-301.
K.In vitro detoxification of aflatoxin B1,deoxynivalenol,fumonisins,T-2 toxin and zearalenone by probiotic bacteria from genus Lactobacillus and Saccharomyces cerevisiae yeast[J].Probiotics and Antimicrobial Proteins,2020,12(1):289-301.
[52] HERNANDEZ -MENDOZA A,GARCIA H S,STEELE J L.Screening of Lactobacillus casei strains for their ability to bind aflatoxin B1[J].Food and Chemical Toxicology,2009,47(6):1064-1068.
[53] HASHEMI S M B,AMIRI M J.A comparative adsorption study of aflatoxin B1 and aflatoxin G1 in almond butter fermented by Lactobacillus fermentum and Lactobacillus delbrueckii subsp.lactis[J].LWT,2020,128:109500.
[54] BEJAOUI H,MATHIEU F,TAILLANDIER P,et al.Ochratoxin A removal in synthetic and natural grape juices by selected oenological Saccharomyces strains[J].Journal of Applied Microbiology,2004,97(5):1038-1044.
[55] BEJAOUI H,MATHIEU F,TAILLANDIER P,et al.Biodegradation of ochratoxin A by Aspergillus section Nigri species isolated from French grapes:A potential means of ochratoxin A decontamination in grape juices and musts[J].FEMS Microbiology Letters,2006,255(2):203-208.
[56] YUE T L,DONG Q F,GUO C X,et al.Reducing patulin contamination in apple juice by using inactive yeast[J].Journal of Food Protection,2011,74(1):149-153.
[57] GONÇALVES B L,ROSIM R E,DE OLIVEIRA C A F,et al.The in vitro ability of different Saccharomyces cerevisiae -Based products to bind aflatoxin B1[J].Food Control,2015,47:298-300.
[58] VAR I,ERGINKAYA Z,KABAK B.Reduction of ochratoxin A levels in white wine by yeast treatments[J].Journal of the Institute of Brewing,2009,115(1):30-34.
[59] LUO Y,WANG J G,LIU B,et al.Effect of yeast cell morphology,cell wall physical structure and chemical composition on patulin adsorption[J].PLoS One,2015,10(8):e0136045.
[60] ![]() A,et al.Toxin-producing fungi on feed grains and application of yeasts for their detoxification[J].Polish Journal of Veterinary Sciences,2013,16(2):391-393.
A,et al.Toxin-producing fungi on feed grains and application of yeasts for their detoxification[J].Polish Journal of Veterinary Sciences,2013,16(2):391-393.
[61] PEREYRA C M,GIL S,CRISTOFOLINI A,et al.The production of yeast cell wall using an agroindustrial waste influences the wall thickness and is implicated on the aflatoxin B1 adsorption process[J].Food Research International,2018,111:306-313.
[62] KOLLÁR R,REINHOLD B B,PETRÁKOVÁ E,et al.Architecture of the yeast cell wall β(1→6)-glucan interconnects mannoprotein,β(1 →3)-glucan,and chitin[J].Journal of Biological Chemistry,1997,272(28):17762-17775.
[63] RAJU M V L N,DEVEGOWDA G.Influence of esterified -glucomannan on performance and organ morphology,serum biochemistry and haematology in broilers exposed to individual and combined mycotoxicosis(aflatoxin,ochratoxin and T-2 toxin)[J].British poultry science,2000,41(5):640-650.
[64] SANTOS A,MARQUINA D,BARROSO J,et al.(1→6)-β-D-glucan as the cell wall binding site for Debaryomyces hansenii killer toxin[J].Letters in Applied Microbiology,2002,34(2):95-99.
[65] YIANNIKOURIS A,FRANCOIS J,POUGHON L,et al.Alkali extraction of β-D-glucans from Saccharomyces cerevisiae cell wall and study of their adsorptive properties toward zearalenone[J].Journal of Agricultural and Food Chemistry,2004,52(11):3666-3673.
[66] LUO Y,LIU X J,LIU Y,et al.Exogenous calcium ions enhance patulin adsorption capability of Saccharomyces cerevisiae[J].Journal of Food Protection,2019,82(8):1390-1397.
[67] SUN X L,HE X X,SIYU XUE K,et al.Biological detoxification of zearalenone by Aspergillus niger strain FS10[J].Food and Chemical Toxicology,2014,72:76-82.
[68] YUAN Y,WANG X,HATAB S,et al.Patulin reduction in apple juice by inactivated A licyclobacillus spp[J].Letters in Applied Microbiology,2014,59(6):604-609.
[69] NIDERKORN V,BOUDRA H,MORGAVI D P.Binding of Fusarium mycotoxins by fermentative bacteria in vitro[J].Journal of Applied Microbiology,2006,101(4):849-856.
[70] TOPCU A,BULAT T,WISHAH R,et al.Detoxification of aflatoxin B1 and patulin by Enterococcus faecium strains[J].International Journal of Food Microbiology,2010,139(3):202-205.
[71] HSU T C,YI P J,LEE T Y,et al.Probiotic characteristics and zearalenone-removal ability of a Bacillus licheniformis strain[J].PLoS One,2018,13(4):e0194866.
[72] BATA Á,LÁSZTITY R.Detoxification of mycotoxin-contaminated food and feed by microorganisms[J].Trends in Food Science &Technology,1999,10(6/7):223-228.
[73] XIA X S,ZHANG Y,LI M Y,et al.Isolation and characterization of a Bacillus subtilis strain with aflatoxin B1 biodegradation capability[J].Food Control,2017,75:92-98.
[74] CIEGLER A,LILLEHOJ E B,PETERSON R E,et al.Microbial detoxification of aflatoxin[J].Applied Microbiology,1966,14(6):934-939.
[75] CSERHÁTI M,KRISZT B,KRIFATON C,et al.Mycotoxin-degradation profile of Rhodococcus strains[J].International Journal of Food Microbiology,2013,166(1):176-185.
[76] ESHELLI M,HARVEY L,EDRADA-EBEL R A,et al.Metabolomics of the bio-degradation process of aflatoxin B1 by actinomycetes at an initial pH of 6.0[J].Toxins,2015,7(2):439-456.
[77] HARKAI P,SZABÓ I,CSERHÁTI M,et al.Biodegradation of aflatoxin-B1 and zearalenone by Streptomyces sp.collection[J].International Biodeterioration &Biodegradation,2016,108:48-56.
[78] ARAI T,ITO T,KOYAMA Y.Antimicrobial activity of aflatoxins[J].Journal of Bacteriology,1967,93(1):59-64.
[79] GUAN S,JI C,ZHOU T,et al.Aflatoxin B1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium[J].International Journal of Molecular Sciences,2008,9(8):1489-1503.
[80] RODRIGUEZ H,REVERON I,DORIA F,et al.Degradation of ochratoxin A by Brevibacterium species[J].Journal of Agricultural and Food Chemistry,2011,59(19):10755-10760.
[81] TENIOLA O D,ADDO P A,BROST I M,et al.Degradation of aflatoxin B1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp.nov.DSM44556T[J].International Journal of Food Microbiology,2005,105(2):111-117.
[82] TAYLOR M C,JACKSON C J,TATTERSALL D B,et al.Identification and characterization of two families of F420H2‐dependent reductases from Mycobacteria that catalyse aflatoxin degradation [J].Molecular Microbiology,2010,78(3):561-575.
[83] RISA A,KRIFATON C,KUKOLYA J,et al.Aflatoxin B1 and zearalenone -detoxifying profile of Rhodococcus type strains[J].Current Microbiology,2018,75(7):907-917.
[84] PRETTL Z,DÉSI E,LEPOSSA A,et al.Biological degradation of aflatoxin B1 by a Rhodococcus pyridinivorans strain in by-product of bioethanol[J].Animal Feed Science &Technology,2017,224:104-114.
[85] SAKAI M,MIYAUCHI K,KATO N,et al.2-Hydroxypenta-2,4-dienoate metabolic pathway genes in a strong polychlorinated biphenyl degrader,Rhodococcus sp.strain RHA1[J].Applied and Environmental Microbiology,2003,69(1):427-433.
[86] LARKIN M J,KULAKOV L A,ALLEN C C R.Biodegradation and Rhodococcus-masters of catabolic versatility [J].Current Opinion in Biotechnology,2005,16(3):282-290.
[87] MARTÍNKOVÁ L,UHNÁKOVÁ B,PÁTEK M,et al.Biodegradation potential of the genus Rhodococcus[J].Environment International,2009,35(1):162-177.
[88] TAHEUR F B,KOUIDHI B,AL QURASHI Y M A,et al.Biotechnology of mycotoxins detoxification using microorganisms and enzymes[J].Toxicon,2019,160:12-22.
[89] RAO K R,VIPIN A V,HARIPRASAD P,et al.Biological detoxification of Aflatoxin B1 by Bacillus licheniformis CFR1[J].Food Control,2017,71:234-241.
[90] PETCHKONGKAEW A,TAILLANDIER P,GASALUCK P,et al.Isolation of Bacillus spp.from Thai fermented soybean(Thua‐nao):Screening for aflatoxin B1 and ochratoxin A detoxification[J].Journal of Applied Microbiology,2008,104(5):1495-1502.
[91] YI P J,PAI C K,LIU J R.Isolation and characterization of a Bacillus licheniformis strain capable of degrading zearalenone[J].World Journal of Microbiology and Biotechnology,2011,27(5):1035-1043.
[92] WANG S W,HOU Q Q,GUO Q Q,et al.Isolation and characterization of a deoxynivalenol-degrading bacterium Bacillus licheniformis YB9 with the capability of modulating intestinal microbial flora of mice[J].Toxins,2020,12(3):184-196.
[93] FARZANEH M,SHI Z Q,GHASSEMPOUR A,et al.Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran[J].Food control,2012,23(1):100-106.
[94] GAO X,MA Q G,ZHAO L H,et al.Isolation of Bacillus subtilis:Screening for aflatoxins B1,M1,and G1 detoxification[J].European Food Research and Technology,2011,232(6):957-962.
[95] WATANAKIJ N,VISESSANGUAN W,PETCHKONGKAEW A.Aflatoxin B1 -degrading activity from Bacillus subtilis BCC 42005 isolated from fermented cereal products[J].Food Additives &Contaminants:Part A,2020,37(9):1579-1589.
[96] CHO K J,KANG J S,CHO W T,et al.In vitro degradation of zearalenone by Bacillus subtilis [J].Biotechnology Letters,2010,32(12):1921-1924.
[97] JU J,TINYIRO S E,YAO W R,et al.The ability of Bacillus subtilis and Bacillus natto to degrade zearalenone and its application in food[J].Journal of Food Processing and Preservation,2019,43(10):e14122.
[98] TINYIRO S E,YAO W R,SUN X L,et al.Scavenging of zearalenone by Bacillus strains in vitro[J].Research Journal of Microbiology,2011,6(3):304-309.
[99] WANG G,YU M Z,DONG F,et al.Esterase activity inspired selection and characterization of zearalenone degrading bacteria Bacillus pumilus ES-21[J].Food Control,2017,77:57-64.
[100] WANG Y,ZHANG J,WANG Y L,et al.Isolation and characterization of the Bacillus cereus BC7 strain,which is capable of zearalenone removal and intestinal flora modulation in mice[J].Toxicon,2018,155:9-20.
[101] CHANG X J,WU Z D,WU S L,et al.Degradation of ochratoxin A by Bacillus amyloliquefaciens ASAG1[J].Food Additives &Contaminants:Part A,2015,32(4):564-571.
[102] SHANG L L,BAI X X,CHEN C,et al.Isolation and identification of a Bacillus megaterium strain with ochratoxin A removal ability and antifungal activity[J].Food Control,2019,106:106743.
[103] WANG J Q,YANG F,YANG P L,et al.Microbial reduction of zearalenone by a new isolated Lysinibacillus sp.ZJ-2016-1[J].World Mycotoxin Journal,2018,11(4):571-578.
[104] KULKARNI M,CHAUDHARI A.Biodegradation of p-nitrophenol by P.putida[J].Bioresource Technology,2006,97(8):982-988.
[105] SAMUEL M S,SIVARAMAKRISHNA A,MEHTA A.Degradation and detoxification of aflatoxin B1 by Pseudomonas putida[J].International Biodeterioration&Biodegradation,2014,86:202-209.
[106] SANGARE L,ZHAO Y,FOLLY Y M E,et al.Aflatoxin B1 degradation by a Pseudomonas strain[J].Toxins,2014,6(10):3028-3040.
[107] TAN H,HU Y C,HE J,et al.Zear alenone degradation by two Pseudomonas strains from soil[J].Mycotoxin Research,2014,30(4):191-196.
[108] TAN H,ZHANG Z M,HU Y C,et al.Isolation and characterization of Pseudomonas otitidis TH-N1 capable of degrading Zearalenone[J].Food Control,2015,47:285-290.
[109] ADEBO O A,NJOBEH P B,SIDU S,et al.Aflatoxin B1 degradation by liquid cultures and lysates of three bacterial strains[J].International Journal of Food Microbiology,2016,233:11-19.
[110] SINGH J,MEHTA A.Protein-mediated degradation of aflatoxin B1 by Pseudomonas putida[J].Brazilian Journal of Microbiology,2019,50(4):1031-1039.
[111] SONG J J,ZHANG S J,XIE Y L,et al.Purification and characteristics of an aflatoxin B1 degradation enzyme isolated from Pseudomonas aeruginosa[J].FEMS Microbiology Letters,2019,366(5):fnz034.
[112] CASTORIA R,MANNINA L,DURÁN-PATRÓN R,et al.Conversion of the mycotoxin patulin to the less toxic desoxypatulinic acid by the biocontrol yeast Rhodosporidium kratochvilovae strain LS11[J].Journal of Agricultural and Food Chemistry,2011,59(21):11571-11578.
[113] LI X H,TANG H,YANG C,et al.Detoxification of mycotoxin patulin by the yeast Rhodotorula mucilaginosa[J].Food Control,2019,96:47-52.
[114] DONG X Y,JIANG W,LI C S,et al.Patulin biodegradation by marine yeast Kodameae ohmeri[J].Food Additives &Contaminants:Part A,2015,32(3):352-360.
[115] ZHENG X F,LI Y L,ZHANG H Y,et al.Identification and toxicological analysis of products of patulin degradation by Pichia caribbica[J].Biological Control,2018,123:127-136.
[116] CAO J,ZHANG H Y,YANG Q Y,et al.Efficacy of Pichia caribbica in controlling blue mold rot and patulin degradation in apples[J].International Journal of Food Microbiology,2013,162(2):167-173.
[117] ZHENG X F,YANG Q Y,ZHANG H Y,et al.The possible mechanisms involved in degradation of patulin by Pichia caribbica [J].Toxins,2016,8(10):289-305.
[118] DIAO E J,HOU H X,HU W C,et al.Removing and detoxifying methods of patulin:A review [J].Trends in Food Science &Technology,2018,81:139-145.
[119] HACKBART H C S,MACHADO A R,CHRISTRIBEIRO A,et al.Reduction of aflatoxins by Rhizopus oryzae and Trichoderma reesei[J].Mycotoxin Research,2014,30(3):141-149.
[120] XING F G,WANG L M,LIU X,et al.Aflatoxin B1 inhibition in Aspergillus flavus by Aspergillus niger through down-regulating expression of major biosynthetic genes and AFB1 degradation by atoxigenic A.flavus[J].International Journal of Food Microbiology,2017,256:1-10.